This sample Salmonella Research Paper is published for educational and informational purposes only. If you need help writing your assignment, please use our research paper writing service and buy a paper on any topic at affordable price. Also check our tips on how to write a research paper, see the lists of health research paper topics, and browse research paper examples.
Introduction
First Identification
In 1886 Daniel E. Salmon, a veterinary pathologist at the U.S. Department of Agriculture, and his coworker Theobald Smith, published a seminal paper describing the isolation of a motile Gram-negative bacillus from a number of cases of swine cholera. The organism was named Bacillus cholerae-suis. In 1900 the generic name of salmonella was proposed to honor Salmon’s many achievements and the organism he first discovered is now known as Salmonella enterica serovar Cholerae-suis.
The Organism
Salmonellae are Gram-negative rods. They are generally motile with pertrichious fimbriae, grow on nutrient agar, do not ferment lactose, and are aero-anaerobes (Threlfall, 2005b). The genus Salmonella is now regarded as comprising two species, Salmonella enterica and Salmonella bongori, with the type species divided into six subspecies listed as follows: Salmonella enterica subsp. enterica (subspecies I), salamae (subspecies II), arizonae (IIIa), diarizonae (IIIb), houtenae (IV), bongori (V), and indica (VI). Within these subspecies over 2400 different serovars, as classified by the Kauffmann and White scheme, have been identified (Poppoff et al., 2004). Of these, the great majority of serovars that cause disease in warm-blooded animals (ca. 99%) fall within Salmonella enterica subspecies I (S. enterica subsp. enterica). For ease of reference in this research paper, such serovars will be referred to as, for example, Salmonella Enteritidis, S. Montevideo, S. Typhimurium, etc. Members of the other five subspecies (II–VI) are, in the main, parasites of cold-blooded animals or are found in the natural environment. Strains of Salmonella arizonae and Salmonella diarizonae have been identified in sheep in several countries, but infections in humans are relatively rare.
Salmonella organisms are found in both the environment and in a wide range of animals. The primary hosts of organisms causing disease in humans are food-producing animals – poultry, cattle, and swine – and the organisms are transmitted to humans either by direct contact or through the medium of infected food. Some serovars are commonly found in cold-blooded animals – for example, reptiles and terrapins, and in such cases infection is normally by direct contact or by contamination of water supplies.
The Disease
In animals raised for consumption, salmonella infection can be either symptomatic or asymptomatic. For example, in cattle, and particularly calves, infection with S. Typhimurium can cause serious systemic disease in addition to severe gastroenteritis, often resulting in death if not treated. Infection with S. Dublin can cause abortions in cattle; in infected calves, it causes severe diarrhea whereas in adult cattle it is responsible for septic abortion associated with a 70% mortality and prolonged carriage by surviving animals. Similarly, in poultry S. Pullorum or Gallinarum can produce severe symptoms characteristic of ‘fowl typhoid.’ In contrast S. Enteritidis in poultry is for the most part asymptomatic, as is S. Derby in pigs.
From a clinical perspective salmonellosis in humans falls into three broad categories: enteric fever, invasive disease (nontyphoidal), and gastroenteritis.
Enteric Fever
Enteric fever, commonly known as typhoid fever, is normally regarded as a disease presentation as a consequence of infection with S. Typhi, which is a host-adapted serovar; infections are restricted to human beings and to closely related anthropoids. The symptoms of typhoid fever in cases of human infection are highly variable with regard to severity and localized consequence. Following ingestion the organisms proceed to the intestinal tract and breach the intestinal wall to reach the lamina propia, where they may establish a local infection or disseminate to establish a systemic infection. Typhoid fever is characterized by prolonged fever, bacterial presence in the reticulo-endethelial section, and significant inflammation of the lymphoid organs of the small intestine. A small number of infected individuals may develop a more severe or complicated disease, including perforation within the small intestine leading to peritonitis, intestinal hemorrhage, myocarditis, encephalopathy, delirium, and meningitis. Chronic carriage of the organism also occurs in some patients, resulting in both relapse and the development of a long-term carrier state. Clinical complications have been associated with factors such as the age of the patient, use of antibiotics, and geographical location. Enteric fever may also develop following infections with S. Paratyphi A and S. Paratyphi B. Although symptoms may be similar to those experienced following infection with S. Typhi, in general these are less severe than those associated with true typhoid. Nevertheless, paratyphoid fever resulting from infection with S. Paratyphi A is increasing in incidence in several countries in South-East Asia and the Indian subcontinent, and there is considerable concern about the long-term implications of this development (see the section ‘Typhoid and paratyphoid fever’).
When the manifestations of systemic disease are mainly septicemic, as is usually the case with typhoid and paratyphoid bacilli in humans, the clinical picture is one of enteric fever with an incubation period of 10–20 days, but with outside limits of 3 and 56 days depending on the infecting dose. Diarrhea, starting 3–4 days after onset of fever and lasting, on average, 6 days, may occur in 50% of cases of typhoid fever and is more common in younger, rather than in older, children or adults; intestinal symptoms, however, may be absent or insignificant.
Invasive Disease (Nontyphoidal)
Certain other serovars – for example, Blegdam, Bredeney, Cholerae-suis, Dublin, and Virchow – are also invasive but tend to cause pyemic infections and to localize in the viscera, meninges, bones, joints, and serous cavities. In developed countries many other serovars may also be invasive in certain circumstances, often related to the infective dose and the host. For instance, invasive disease by serovars generally regarded as noninvasive frequently occurs in immunocompromised patients or in patients with other underlying diseases. In developing countries a different picture has emerged, with serovars normally regarded as noninvasive – for example, Typhimurium, Wien, and Senftenberg being associated with highly virulent infections with a high degree of morbidity and mortality. Such infections have been associated with the carriage of plasmids that may enhance the invasive potential of their host organism (see the section ‘Developing countries’ under ‘Nontyphoidal salmonellas’).
Gastroenteritis
The most common symptoms following infection with the ubiquitous nontyphoidal serovars found in a number of animal species are those of an acute but mild to moderate enteritis with a short incubation period of 12–48 hours, occasionally as long as 4 days. As with the majority of gastrointestinal bacterial pathogens, symptoms can be more severe in vulnerable patient groups such as young children, the elderly, debilitated, and immunocompromised patients.
Treatment
For enteric fever, including infections with S. Typhi and S. Paratyphi A and B, treatment with an appropriate antibiotic is essential and can be life-saving. As treatment may commence before the results of antimicrobial sensitivities are known it is important to be aware of options and possible problems before beginning treatment (see the section ‘Antimicrobial drug resistance’). The same strictures apply for invasive infections with nontyphoidal salmonellae. Multiple resistance, often including resistance to ‘critical’ antimicrobials, is becoming increasingly common for certain serovars and although not considered important for uncomplicated gastroenteritis, can be very important should extraintestinal spread occur. For uncomplicated gastroenteritis rehydration therapy is considered appropriate in most cases.
Epidemiology
On a global scale it has been estimated that salmonella is responsible for an estimated 3 billion human infections each year. The World Health Organization (WHO) has estimated that typhoid fever accounts for 22 million of these cases and is responsible for 200 000 deaths annually (Crump et al., 2004).
Typhoid And Paratyphoid Fever
Typhoid fever is a significant cause of morbidity and mortality among children and adults in developing countries. The organism remains endemic in developing countries in Africa, South and Central America, and the Indian subcontinent. The disease is also commonly reported from the Middle East, and some countries in Southern and Eastern Europe. In contrast, in developed countries such as the UK or the United States, the incidence of S. Typhi is much lower, with the majority of cases in travelers returning from endemic areas. For example, in the UK, between 150 and 300 cases occur each year with at least 70% of cases in patients with a history of recent foreign travel. Similarly, in the United States, 293 infections were reported in the 12-month period from 1 June 1996 to 31 May 1997, of which 81% were recorded in patients with a history of recent travel to endemic areas.
The epidemiology of paratyphoid fever is less well documented than that of typhoid. Nevertheless, the WHO has estimated that up to 25% of enteric fevers may be caused by S. Paratyphi A and the disease is becoming increasingly common in several countries in South-East Asia, including Vietnam, India, and Nepal.
Nontyphoidal Salmonellas
Data on infections caused by nontyphoidal salmonellas are difficult to quantify, mainly because of differences in surveillance systems or, indeed, the complete lack of such systems in many countries.
Developed countries
Within the European Union (EU) studies by the Enternet surveillance network have provided data for about 150 000 human infections annually, with approximately 1000 deaths, from figures submitted by the 22 countries that report into Enter-net. In the United States, it has been estimated that there are approximately 200 000 infections annually.
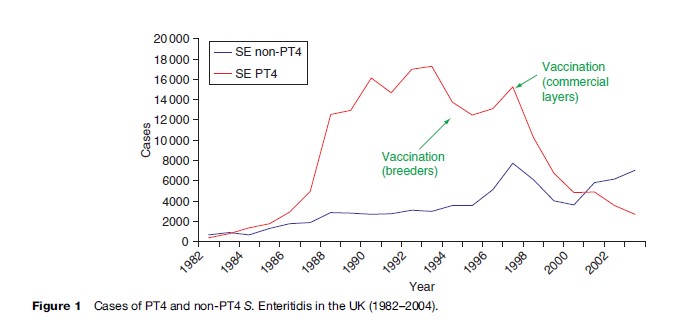
For the last two decades the most common serovar in cases of infection with nontyphoidal salmonellas in the EU has been S. Enteritidis (Saheed et al., 1999). This serovar has its reservoir in poultry, and from 1987 to 2000 the most common phage type (PT) within the serovar was PT 4.
From 1987 to 2000 it was estimated that PT 4 was responsible for over 500 000 cases of S. Enteritidis in the United Kingdom alone (Figure 1). S. Enteritidis PT 4 was unusual in that the organism was transmitted vertically through the oviduct of infected birds, and many infections in humans were traced to infected eggs. Outbreaks were compounded by improper cooking techniques and by the storage at ambient temperatures of dishes made from raw eggs. Despite instructions on the cooking and handling of eggs, the outbreak was only contained by the vaccination of poultry flocks, namely breeding flocks in 1994, and commercial layers in 1998. Regrettably, since 2000, a considerable number of infections caused by non-PT 4 strains have been increasingly identified in the UK. A well-documented series of outbreak investigations and laboratory studies, supplemented by surveys of eggs on retail sale, have traced the source of these strains to eggs imported into the UK from Spain. As a result of various actions, including educating restauranteurs and also informing appropriate authorities within Spain, infections with these non-PT 4 strains have shown a marked decline since 2004.
In the United States, similar problems with S. Enteritidis have been encountered, traced back to contaminated egg dishes. In contrast to the United Kingdom, the predominant PTs were 8 and 13a, although outbreaks of PT 4 not associated with foreign travel have been recognized since 1993.
Other serovars with an international distribution in developed countries have included S. Typhimurium definitive phage type (DT) 104. This organism exhibits chromosomally mediated multiple resistance (MR) to five antimicrobials – ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracyclines (R-type ACSSuT). MR S. Typhimurium DT 104 of R-type ACSSuT was first identified in the UK in the early 1980s from gulls and exotic birds. With the exception of a small outbreak in Scotland in the mid-1980s there were no isolations from humans until 1989, by which time MR S. Typhimurium DT 104 had also been isolated from cattle. Over the next 5 years this strain became epidemic in bovine animals throughout the UK, and also in poultry, particularly turkeys, and in pigs and sheep. Human infection with MR S. Typhimurium DT 104 has been associated with the consumption of chicken, beef, pork sausages, and meat paste and to a lesser extent with occupational contact with infected animals. This particular clone has subsequently caused outbreaks of infection in food animals and humans in numerous European countries and as far afield as South Africa, the United Arab Emirates, and the Philippines (Threlfall, 2000). In 1996 infections with MR DT 104 were recognized in cattle and humans in North America, both in Canada and in the United States. Of particular concern has been the resistance of the organism to a wide range of therapeutic antimicrobials. Furthermore, in some countries there have been reports of an apparent predilection of the organism to cause serious disease, although this is not the case in the UK.
In the United States, closely related strains of MR S. Newport with plasmid-encoded resistance to ceftriaxone have been associated with numerous infections in both cattle and humans. This has caused it to become the third most common serotype causing salmonellosis in man in the United States from 2000 to 2002. The organism commonly shows resistance to ACSSuT with additional resistance to third-generation cephalosporins mediated by the CMY-2 beta-lactamase gene (see ‘Developed countries’ under ‘Other salmonella serovars’).
Because of the association of many nontyphoidal salmonella serovars with food-production animals, many outbreaks have been linked to foods of animal origin or to foods contaminated with animal waste. This, coupled with the massive importation of foods between countries, including both developed and developing countries, has ensured that products contaminated with salmonella organisms have been widely distributed. An enormous diversity of vehicles of infection have been identified, ranging from salad vegetables to spices to coconut, as well as traditional animal-associated products such as poultry and poultry products, milk, cheese, and undercooked beef and pork. The EU-funded Enter-net surveillance network has identified over 50 such food-borne salmonella outbreaks since its inception in 1992 (Fisher and Threlfall, 2005; Cook et al., 2006), and, in many cases, has been instrumental in the development of intervention measures that have either contained existing outbreaks or have resulted in the removal of contaminated products from the food chain.
A further significant source of salmonellosis in developed countries is pet reptiles. Many reptiles carry salmonella organisms as part of their normal bacterial flora and, although the types of salmonella in reptiles are not those normally associated with large outbreaks, such as those caused by S. Enteritidis, the reptile-associated types can cause very serious illness, particularly in vulnerable patient groups such as young children or the elderly. There have been many cases of reptile-associated salmonellosis documented worldwide. In the United States in the 1970s, a large outbreak involving several thousand people was attributed to turtles; as a result, turtle hatching was banned in the United States in 1975.
Developing Countries
Since 1970, nontyphoidal salmonellas of several different serovars have caused extensive outbreaks in many developing countries. The common pattern has been for several hospitals, often situated many miles apart, to be involved. The majority of outbreaks have occurred in neonatal and pediatric wards, but community outbreaks in villages and small towns have also been reported. The clinical disease has been severe with enteritis frequently accompanied by septicemia and, in several outbreaks, a mortality rate of up to 30% has been reported. Serotypes involved include S. Typhimurium in the Middle East and the Indian subcontinent, and S. Wien in southern Europe, North Africa, and India, although infections caused by other serovars have been reported, notably S. Senftenberg in India and S. Johannesburg in South Africa. A common feature of all strains has been resistance of up to 10 antimicrobial drugs (see Rowe and Threlfall, 1984; Threlfall, 2005a for extensive discussion of such outbreaks and the features of strains involved).
A particular feature of outbreaks of infections with MR, nontyphoidal salmonellas in developing countries has been the lack of involvement of food animal reservoirs. Spread has for the most part been by person-to-person contact and antibiotic resistance has developed as a result of the use of antibiotics in human medicine, particularly in those countries where there is little control over the use of antibiotics. The most common presentation has been severe enteritis and cases of septicemia have also been reported, with high mortality in many outbreaks.
Identification And Typing
Serotyping
Strains of Salmonella spp. are classified into serovars on the basis of extensive diversity of the heat-stable lipopolysaccharide (O) antigens and heat-labile flagellar protein (H) antigens in accordance with the scheme instituted by White (1926), and extended and elaborated by Kauffmann (1972). The resultant Kauffmann and White scheme (Popoff et al., 2004) is recognized worldwide and remains the definitive method for the serological identification of salmonellae.
Subdivision Within Serovars: Phenotypic Subtyping
A variety of phenotypic methods have been used both independently and in combination for subdivision within serovars. Those currently in use include bacteriophage typing (phage typing) and resistance (antibiogram) typing.
Bacteriophage Typing
The underlying principle of phage typing is the host specificity of bacteriophages and on this basis several phage-typing schemes have been developed for serovars of clinical or epidemiological importance. (For extensive review of the strengths and weaknesses of these schemes, see White, 1926; Threlfall, 2005a.) The most important schemes internationally are those for S. Typhi, S. Paratyphi A and B, S. Enteritidis, S. Typhimurium, and S. Virchow.
Salmonella Typhi
The first phage typing scheme was based on the principle of phage adaptation and was developed for the differentiation of Typhi; in this scheme, progressive adaptations were made of Vi phage II – specific for Vi (capsular) antigen of Typhi – which is highly adaptable and shows a high degree of specificity for the last strain on which it has been propagated. The extraordinary adaptation of Vi phage II is due in part to the selection of spontaneously occurring host-range phage mutants by the bacterium and in part to a nonmutational phenotypic modification of phage by the host strain. The method of Vi phage typing was standardized in 1947 and with further adaptations of Vi phage II, a further 95 types have been defined and internationally recognized, bringing the total number of Vi PTs to 106.
Other Serovars
In contrast to the phage typing scheme for Typhi, phage typing schemes for other serovars depend on, to a limited extent, phage adaptability and, for the most part, are based on patterns of lysis produced by serologically distinct phages isolated from a variety of sources. More than 70 PTs are now recognized in the Enteritidis scheme, the value of which was realized on an international scale following the global pandemic of Enteritidis from the late 1980s, extending to 2006. This scheme was instrumental in identifying the global pandemic PT 4, and more recently, in detecting and monitoring the emergence of non-PT 4s associated with eggs from poultry flocks in several different European countries. A major achievement has been the standardization of phage typing for S. Enteritidis throughout Europe and one scheme, that of Ward and colleagues (1987), is now in use in reference laboratories for human salmonellosis in 22 European countries as well as in Australia, Japan, and Canada (Fisher and Threlfall, 2005).
For S. Typhimurium, almost 300 PTs have now been recognized and designated using the scheme of Anderson and colleagues (1977). This scheme is the most commonly used worldwide, although local schemes have been developed for individual countries. The importance of this scheme is well illustrated by the universal recognition of the multiple drug-resistant (MDR) epidemic clone of S. Typhimurium DT 104. However, of note is that PTs cannot always be regarded as indicative of clonality because PT conversions may result from the acquisition of both plasmids and bacteriophages. A further problem with phage typing is that because of the necessity to propagate and maintain bacteriophage stocks and to implement strict quality control procedures, the procedure is best performed by highly trained staff in reference laboratories and not by workers in individual laboratories on an ad hoc basis.
Resistance (Antibiogram) Typing
The pattern of susceptibility/resistance to selected antimicrobial drugs can be a very useful screen for epidemiological investigations. Because of mutation and/or plasmid acquisition such patterns cannot be regarded as definitive. Nevertheless, patterns such as ACSSuT for S. Typhimurium DT 104 and for the identification of Salmonella Genomic Island 1 (SGI-1), and ACSSuTTm (Tm, trimethoprim) for S. Typhi have become very useful markers and have been used on a global basis to assist in the identification of epidemic clones or drug resistance islands.
Of particular importance in the use of susceptibility/ resistance patterns (R-types) as epidemiological markers, and also in the international surveillance of antimicrobial drug resistance in salmonella is the standardization/harmonization of methodologies coupled with the interpretation of results. For human salmonella isolates within Europe this has been achieved, in the first instance, by the harmonization of methods of susceptibility testing in all reference laboratories with responsibility for human referrals. Following international agreement on the definitions of resistance and susceptibility based on various methodologies, it has been possible to combine and analyze the antimicrobial drug resistance data originating from all countries within Europe who participate in the Enter-net surveillance network.
Subdivision Within Serovars: Molecular Subtyping
A range of molecular biological tools based on characterization of the genotype of the organism by analysis of plasmid and chromosomal DNA have now been developed either to supplement the more traditional phenotypic methods of typing (serotyping, phage typing, biotyping) or, in some cases, as methods of discrimination in their own right (see Threlfall, 2005b; Cook et al., 2006).
Plasmid Typing
Many strains of salmonellae carry plasmids differing in both molecular mass and number. Plasmid typing based on the numbers and molecular mass of plasmids after extraction of partially purified plasmid DNA has been used for differentiation within serovars. Plasmid typing is therefore restricted to serovars possessing plasmids and is of limited use in those serovars in which the majority of isolates contain only one plasmid, or are plasmid-free. The sensitivity of the plasmid profile typing may be increased by cleaving plasmid DNA with a limited number of restriction endonucleases and the resultant plasmid ‘fingerprint’ may be used to discriminate between plasmids of similar molecular mass. More recently the characterization of plasmids by identification of specific replicon areas has added a new dimension to plasmid typing. This method, developed by Caratollli and colleagues (2005), has considerable potential not only for the identification of plasmid incompatibility groups but also for investigating the spread of such plasmids, and the resistance genes encoded thereon, through the food chain.
Identification Of Chromosomal Heterogeneity
Molecular typing methods based on the characterization of plasmid DNA include plasmid profile typing, plasmid fingerprinting, and the identification of plasmidmediated virulence genes. Chromosomally based methods have sought to identify small regions of heterogeneity within the bacterial chromosome. Of the latter the most commonly used have been ribotyping, insertion sequence (IS) 200 typing, and pulsed field gel electrophoresis (PFGE). The latter method permits analysis of the whole bacterial genome on a single gel and is currently regarded as the gold standard for the molecular subtyping of salmonella. This method is used as the basic method of subtyping of salmonella in the United States, and for subdivision within PTs in those countries which use phage typing as the primary method for the discrimination of epidemiologically important serovars. The method has become standardized and networks have been developed – PulseNet in the United States (Swaminathan et al., 2001) and SalmGene in Europe (Fisher and Threlfall, 2005) – to provide common, harmonized molecular typing methods, and to facilitate the rapid electronic transfer of the images captured by them in a digitized format. More recently the SalmGene database of PFGE types has been expanded to form the basis of PulseNet Europe, which is fully compatible with PulseNet USA and other PulseNet networks, thereby providing an encompassing network for the molecular subtyping of salmonella worldwide.
With the development of the polymerase chain reaction (PCR), methods based on amplification of specific DNA sequences to produce characteristic groups of fragments dependent on the origin of the template DNA have been developed and used, with some success. Such methods include random amplified polymorphic DNA typing (RAPD), enterobacterial repetitive intergenic consensus typing (ERIC-PCR), repetitive extragenic palindromic element typing (REP-PCR), amplified fragment length polymorphism fingerprinting (AFLP), and variable number of tandem repeats (VNTR) fingerprinting. VNTR fingerprinting is based on the presence and subsequent identification of units of repeated DNA elements in the genome. Such elements, known as VNTRs, range from about 10 to 100 base pairs (bps). VNTR fingerprinting has been applied to the subtyping of S. Typhi and S. Typhimurium. A major drawback of the method is that for meaningful results VNTRs for typing should be based on the published genome sequence of a serovar. As only a limited number of serovar sequences have been published, the applicability of this method is somewhat limited.
Antimicrobial Drug Resistance
Resistance to key antimicrobials is particularly important in the treatment of infections caused by S. Typhi and S. Paratyphi A. The increasing occurrence of multiple resistance in serovars other than Typhi has also had a profound effect in the treatment of salmonella septicemia in infants and young children in developing countries, where multiple resistant strains have been implicated in numerous outbreaks in the community and in hospital pediatric units for the past 30 years.
Salmonella Typhi And Paratyphi A
Salmonella Typhi
An appropriate antibiotic is essential for the treatment of patients with typhoid fever and should commence as soon as clinical diagnosis is made. Since the emergence of plasmid-mediated chloramphenicol resistance in the typhoid bacillus in the early 1970s, the efficacy of chloramphenicol as a first-line drug has been increasingly undermined by outbreaks caused by strains with resistance to this antimicrobial in countries as far apart as Mexico and India. A feature of chloramphenicol-resistant strains from such outbreaks was that although the strains belonged to different Vi PTs, resistance to chloramphenicol – often in combination with resistance to streptomycin, sulfonamides, and tetracyclines (R-type CSSuT) – was encoded by a plasmid of the H1 incompatibility group (now termed HI1). Since 1989 there have been many outbreaks caused by Typhi strains resistant to chloramphenicol, ampicillin, and trimethoprim, and additional resistances to streptomycin, sulfonamides, and tetracyclines (R-type ACSSuTTm), particularly in the Indian subcontinent (see Threlfall 2005a, b; Cooke et al., 2006). The emergence of strains with resistance to trimethoprim and ampicillin, in addition to chloramphenicol, has caused many treatment problems.
Without exception, in all outbreaks studied thus far involving MR Typhi, the complete spectrum of multiple resistance has been encoded by plasmids of the HI1 incompatibility (inc) group and it has been suggested, incorrectly, that this plasmid group is specific for the typhoid bacillus. Evolutionary diversity within the HI1 group has recently been observed in plasmids from MR strains of S. Typhi from Vietnam over a 10-year period during the 1990s.
Since 1989, following the emergence of strains with resistance to chloramphenicol, ampicillin and trimethoprim, ciprofloxacin (CpL) has become the first-line drug in both developing and developed countries. Regrettably, strains of S. Typhi with decreased susceptibility to CpL (minimal inhibitory concentration (MIC): 0.25–1.0 mg/L) have been increasingly reported. In such strains CpL is chromosomally encoded. Such strains have caused substantive outbreaks in several developing countries, notably Tajikistan and Vietnam, and have also caused treatment problems in developed countries. Azithromycin, a macrolide antibiotic, has also been evaluated for the treatment of infections caused by MR typhoid, with encouraging results.
Salmonella Paratyphi A
Infections caused by S. Paratyphi A may also require antimicrobial intervention. S. Paratyphi A with decreased susceptibility to CpL has been reported in India since the late 1990s. An increase in strains of S. Paratyphi A with decreased susceptibility to CpL from patients from 10 European countries between 1999 and 2001 has also been observed and in the UK in 2005 over 80% of isolates of S. Paratyphi A exhibited decreased susceptibility to CpL.
Other Salmonella Serovars
Developed countries
In developed countries salmonella infections are primarily zoonotic in origin. When resistance is present, it has often been acquired prior to transmission of the organism through the food chain to humans. The most important serovars in the UK and Europe are Enteritidis and Typhimurium and, in the United States, Typhimurium, Enteritidis, and more recently, Newport. For all these serovars the main method of spread is through the food chain. In most cases the clinical presentation is that of mild to moderate enteritis. The disease is usually self-limiting and antimicrobial therapy is seldom required.
Since 1991 there has been an epidemic in cattle and humans in England and Wales of MR strains of S. Typhimurium DT 104 of R-type ACSSuT (see the previous section ‘Developed countries’ under ‘Nontyphoidal salmonellas’).
In MR DT 104 of R-type ACSSuT, resistances are contained in a 16-kilobase (kb) region of the 43-kb SGI-1, made up of integrons containing (respectively) the ASu (blaCARB-2 and sul1) and SSp (aadA2) genes (Sp, spectinomycin), with intervening plasmid-derived genes coding for resistance to chloramphenicol/florphenicol (florR) and tetracyclines (tetG). Although chromosomally encoded in recent years, SGI-1 has been identified in several different salmonella serovars, including S. Agona, S. Albany, and S. Paratyphi B variant Java, which is indicative of phage-mediated transfer of resistance, or transfer by an as yet unidentified method. Such strains have caused infections in humans and cattle and there is speculation of a connection with ornamental fish originating in the Far East.
In the United States, multiple resistance has been reported in serovars Saintpaul, Heidelberg, and Newport, in addition to S. Typhimurium DT 104. More recently, MDR S. Newport with plasmid-encoded resistance to ceftriaxone has caused numerous infections in both cattle and humans in North America (Threlfall, 2005a). This organism commonly shows resistance to ACSSuT, with additional resistance to third-generation cephalosporins mediated by the CMY-2 beta-lactamase gene. Similarly there have been increasing reports of resistance to extended-spectrum beta-lactamases in salmonella from humans and food animals in numerous countries worldwide. For example CTX-M-9, -15, and -17 to -18 enzymes have recently been reported in six different serovars isolated from humans in the UK, and CTX-M-like enzymes have been reported in S. Virchow in Spain and in S. Anatum in Taiwan. In Taiwan a particularly alarming development has been the emergence of a highly virulent strain of S. Cholerae-suis with high-level resistance to Cp and with plasmid-mediated resistance to ceftriaxone. As far as is known, the strain has not spread and no further infections with this highly drug-resistant organism have been detected.
Developing Countries
An additional feature of strains on nontyphoidal salmonellas in developing countries has been the possession of plasmid-mediated MDR, often with resistance to seven or more antimicrobials, mediated by a plasmid of the FI incompatibility group. In addition to coding for multiple resistance, this plasmid also codes for production of the hydroxamate siderophore aerobactin, a known virulence factor for some enteric and urinary tract pathogens. These plasmids, first identified in a strain of S. Typhimurium DT 208 that caused numerous epidemics in many Middle Eastern countries in the 1970s, have subsequently been identified in a strain of S. Wien responsible for a massive epidemic that began in Algiers in 1969 but spread rapidly thereafter through pediatric and nursery populations in many countries throughout North Africa, Western Europe, the Middle East, and eventually the Indian subcontinent over the next 10 years. A retrospective molecular study of this group of plasmids has demonstrated that the plasmids have evolved through sequential acquisition of integrons carrying different arrays of antibiotic resistance genes. Although not clinically proven, the epidemiological evidence strongly suggests that possession of this class of plasmid has contributed to the virulence and epidemicity of such strains.
Virulence Aspects Of Salmonella
Salmonella virulence is a highly complex phenomenon, and has been the subject of many investigations. Some serovars such as Typhi, Pullorum, Gallinarum, Dublin, Cholerae-suis, and Enteritidis are highly host-specific, with their reservoirs of infection being anthropoids (Typhi), poultry (Pullorum, Gallinarum, Enteritidis), swine (Cholerae-suis), and cattle (Dublin). Some of these serovars – for instance, Typhi, Gallinarum, and Pullorum – cause disease for the most part only in their natural reservoir. Others, such as Cholerae-suis and Dublin, cause disease in their food animal reservoir but when infections occur outside of their normal reservoir (e.g., in humans), the symptoms can be very severe, often resulting in septicemia with subsequent mortality. Other host-adapted serovars (e.g., Enteritidis) cause little overt disease in their natural reservoir but when transmitted to humans can be a major cause of salmonellosis. Still other serovars (e.g., Typhimurium) have a wide host range and cause disease both in their food animal reservoir (e.g., cattle) and in humans.
Salmonella Pathogenicity Islands
Many of the major components required by S. enterica to cause infections are chromosomally encoded. The regions responsible for the virulence functions are termed salmonella pathogenicity islands (SPI); 14 such islands have now been identified, termed SPI-1–SPI-14. The size, distribution, and virulence functions of these SPIs have been extensively reviewed by Morgan (2006). It is noteworthy that not all serovars possess all the islands, and that differential pathogenicity in different hosts may be related to the presence or absence of such islands.
Salmonella Enterica Virulence Plasmids
In addition to possessing pathogenicity islands certain serovars of subspecies I harbor serovar-specific plasmids ranging from 40–90 kb, which poses a gene cluster promoting virulence in mice. This gene cluster, termed the salmonella plasmid virulence (spv) cluster has been identified in the epidemiologically important serovars Enteritidis, Typhimurium, Dublin, and Gallinarum. The biology of the spv cluster (Tezcan-Merdol et al., 2006) indicates involvement in serum resistance and invasion, but not in the initial phase of the disease in human salmonellosis.
Control
Control of salmonella disease can be exerted at three levels – the individual, the community (the herd), and the environment. Such control may be exerted by vaccination, by eradication and/or withdrawal of an infected product, and by general hygiene and cooking practices. Factors exacerbating the emergence and spread of particular strains, such as the indiscriminate use of antibiotics, may also be important, and the control of antibiotics, particularly in animal husbandry, has been highlighted as an important factor in combating the emergence of strains with resistance to key antibiotics.
Vaccination
Individuals may be vaccinated and, for the control of typhoid, a range of vaccines are available for Salmonella Typhi. The oral, live attenuated Ty21a vaccine (marketed as Vivotif) remains in use in many countries, particularly in the Indian subcontinent. Possibly the most current is the polysaccharide capsular Vi vaccine (Typhim), but assessment of the relative efficacy of the different vaccines is difficult. Several field trials are currently in progress, both in developing and developed countries, with potentially promising results reported for a Vi-conjugate vaccine.
In the control of S. Enteritidis in poultry in the UK, the development and use of vaccines for breeders and layers has been extremely effective, and the use of such vaccines is currently being assessed for use in other countries, particularly within the EU.
Eradication And Withdrawal
In many outbreaks control has been exerted either by eradication of the reservoir of infection (e.g., infected poultry flocks) or withdrawal of the contaminated food products. There are many examples of the latter method of control, one of the most recent being the withdrawal of contaminated confectionary products worldwide following contamination with S. Montevideo. In such instances the existence of an international rapid response network has proved invaluable (see Fisher and Threlfall, 2005 for other examples of product withdrawal at an international level).
General Hygiene And Cooking Practices
The importance of a clean and safe water supply cannot be overestimated. Contaminated water remains a major reservoir of salmonella organisms in developing countries, and many outbreaks of typhoid and paratyphoid have been linked to sewage contamination. Similarly, in developed countries contaminated salad vegetables have been linked to many outbreaks in recent years, with contamination resulting from the use of untreated water for irrigation. Cooking practices and kitchen hygiene are also of major importance, as has been exemplified on an international scale in relation to the use of raw or lightly cooked eggs for both domestic and institutional catering. It is vitally important to thoroughly cook food whenever possible, to wash ‘ready-to-eat’ foods despite any instructions or implications (e.g., ‘already washed’) to the contrary, and to adopt basic hygiene measures such as hand washing after contact with pets, exotic or otherwise. Awareness of the potential hazards on the part of food producers and the general public can play a large part in reducing the burden of infection imposed by salmonella, a common and potentially lethal pathogen.
Bibliography:
- Anderson ES, Ward LR, De Saxe MJ, and De Sa JDH (1977) Bacteriophage-typing designations of Salmonella typhimurium. Journal of Hygiene 78: 297–300.
- Carattoli A, Bertini A, Lilla L, Falbo V, Hopkins K, and Threlfall EJ (2005) Identification of plasmids by PCR-based replicon typing. Journal of Microbiological Methods 63: 219–228.
- Cook FJ, Threlfall EJ, and Wain J (2006) Current trends in the spread and occurrence of human salmonellosis: Molecular typing and emerging antibiotic resistance. In: Rhen M,
- Maskell D, Mastroeni P and Threlfall EJ (eds.) Salmonella: Molecular Biology and Pathogenesis, pp. 1–29. Norfolk, UK: Horizon Bioscience.
- Crump JA, Luby SP, and Mintz ED (2004) The global burden of typhoid fever. Bulletin of the World Health Organisation 82: 346–353.
- Fisher IST and Threlfall EJ (2005) The Enter-net and Salm-gene databases of food-borne bacterial pathogens causing human infections in Europe and beyond: an international collaboration in the development of intervention strategies. Epidemiology & Infection 133: 1–7.
- Kauffmann F (1972) Serological diagnosis of Salmonella species. Copenhagen, Denmark: Munksgaard.
- Morgan E (2006) Salmonella pathogenicity islands. In: Rhen M, Maskell D, Mastroeni P and Threlfall EJ (eds.) Salmonella: Molecular Biology and Pathogenesis, pp. 67–88. Norfolk, UK: Horizon Bioscience.
- Popoff MY, Bockemuhl J, and Gheesling LL (2004) Supplement 2002 (no. 46) to the Kauffmann-White scheme. Research in Microbiology 155: 568–570.
- Rowe B and Threlfall EJ (1984) Drug resistance in Gram-negative aerobic bacilli. British Medical Bulletin 40: 68–76.
- Saheed AM, Gast RK, Potter ME and Wall PG (eds.) (1999) Salmonella Enterica Serovar Enteritidis in Humans and Animals: Epidemiology, Pathogenesis and Control. Iowa, USA: Iowa State University Press.
- Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV, and PulseNet Task Force (2001) PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance. Emerging Infectious Diseases 7: 382–399.
- Tezcan-Merdol D, Ygberg SE, and Rhen M (2006) The Salmonella enterica virulence plasmid and the spv gene cluster. In Rhen M, Maskell D, Mastroeni P, and Threlfall EJ (eds.) Salmonella: Molecular Biology and Pathogenesis. pp. 89–103. Norfolk, UK: Horizon Bioscience.
- Threlfall EJ (2000) Epidemic Salmonella typhimurium DT 104 – a truly international epidemic clone. Journal of Antimicrobial Chemotherapy 46: 7–10.
- Threlfall EJ (2005a) Antibiotic resistance in Salmonella and Shigella. In: White DG, Alkeshun MN and McDermott P (eds.) Frontiers in Antibiotic Resistance: A tribute to Stuart B. Levy, pp. 367–373. Washington, DC: ASM Publications 2005.
- Threlfall EJ (2005b) Salmonella. In: Borriello SP, Murray PR and Funke G (eds.) Topley and Wilson’s Microbiology and Microbial Infections, 10th Edition, part VI, pp. 1398–1434. London: Hodder Arnold.
- Threlfall EJ and Frost JA (1990) The identification, typing and fingerprinting of Salmonella: laboratory aspects and epidemiological applications. Journal of Applied Bacteriology 68: 5–16.
- Ward LR, De Sa JDH, and Rowe B (1987) A phage-typing scheme for Salmonella enteritids. Epidemiology & Infection 99: 291–294.
- White PB (1926) Further Studies of the Salmonella Group. Great Britain Medical Research Council Special Report, no. 103. London: His Majesty’s Stationery Office.
- Bale JA, De Pinna E, Threlfall EJ, and Ward LR (in press) Salmonella Identification: Serotypes and Antigenic Formula. Kauffmann-White Scheme 2007. London: Health Protection Agency.
- Borriello SP, Murray PR and Funke G (eds.) (2005) Topley and Wilson’s Microbiology and Microbial Infections, 10th edn., vol 2, part 4. London: Hodder Arnold.
- Cook GC and Zumla AI (eds.) (in press) Manson’s Tropical Diseases, 22nd edn. London: Elsevier.
- Rhen M, Maskell D, Mastroeni P, and Threlfall EJ (eds.) (2006) Salmonella: Molecular Biology and Pathogenesis. Norfolk, UK: Horizon Bioscience.
- Saheed AM, Gast RK, Potter ME and Wall PG (eds.) (1999) Salmonella Enterica Serovar Enteritidis in Humans and Animals: Epidemiology, Pathogenesis and Control. Ames, IA: Iowa State University Press.
See also:
Free research papers are not written to satisfy your specific instructions. You can use our professional writing services to buy a custom research paper on any topic and get your high quality paper at affordable price.